- Home
- Research
- Research Cores and Services
- Pharmacokinetic and Mass Spectrometry Core
- PKMS Knowledge Center
- Academic Departments
- Research
- Academic Departments
- Research in Clinical Pharmacy
- Research in Medicinal Chemistry
- Research in Pharmaceutical Sciences
- Research Cores and Services
- Biointerfaces Institute
- Michigan Drug Discovery
- Michigan Institute for Clinical & Health Research
- Translational Oncology Program
- Faculty Publications
- News
- UM Pharmacy Professor Research Outreach (PRO)
- About the College
PKMS Knowledge Center
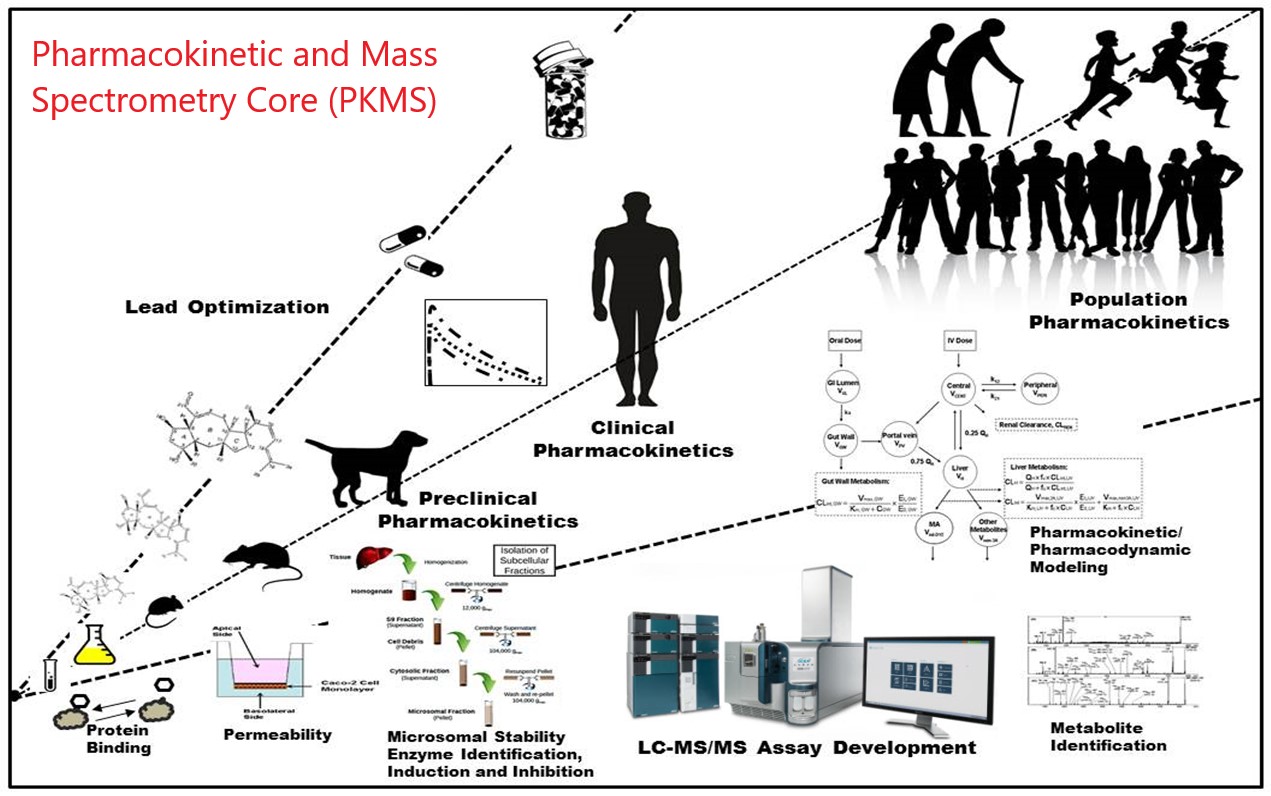
Pharmacokinetics and Mass Spectrometry Core Facility (PKMS) is a shared core resource facility in UM, and serves the scientific research community at UM and beyond with state-of-the-art expertise, methodology, and instrumentation. We are pleased to offer a complete PK service to your research projects with different options including collaboration, Fee-for-Service (FFS) and self-service. Please check our complete PK analysis service - from in vitro and in vivo drug discovery to bioanalysis and Mass spectrometry imaging.
Pharmacokinetics (PK) is the study of the time course of drug’s absorption, distribution, metabolism, and excretion (ADME), what the body interact with a drug, related to the movement of drug into, pass, and out of the body. In order for a drug to be effective in the body, enough of the active form must reach the target to elicit the desired effect. The compound with the best activity against the target enzyme or receptor may have very poor ADME characteristics such as metabolic instability or poor intestinal absorption. When choosing a lead candidate from multiple compounds, the activity of the drug in the efficacy screen must be balanced with acceptable ADME and stability characteristics. Pharmacokinetic studies with mass spectrometry bioanalysis can provide the quantitative information about the half-life of the administrated compound from in vitro and in vivo test, and more useful information of how quickly it can be metabolized or excreted by enzymatically or nonenzymatically. To track the drug accumulation and distribution in organism from in vivo PK study, the mass spectrometry imaging and quantification can come up with innovative solutions to quantify dynamic drug exposures in all targeted tissues, or using artificial intelligence modeling to predict the drug exposure in disease or normal organs.
Representaive Case Study from PKMS Core: https://pharmacy.umich.edu/pkcore/supported-work
1. Borkin D, He SH, Miao HZ, Kempinska K, Pollock J, Chase J, Purohit T, Malik B, Zhao T, Wang JY, Wen B, Zong HL, Jones M, Danet-Desnoyers G, Guzman ML, Talpaz M, Bixby DL, Sun DX, Hess JL, Muntean AG, Maillard I, Cierpicki T, Grembecka J. Pharmacologic Inhibition of the Menin-MLL Interaction Blocks Progression of MLL Leukemia In Vivo. Cancer Cell. 2015;27(4):589-602. PMCID: PMC4415852
2. Li W, Tanikawa T, Kryczek I, Xia H, Li G, Wu K, Wei S, Zhao L, Vatan L, Wen B, Shu P, Sun D, Kleer C, Wicha M, Sabel M, Tao K, Wang G, Zou W. Aerobic Glycolysis Controls Myeloid-Derived Suppressor Cells and Tumor Immunity via a Specific CEBPB Isoform in Triple-Negative Breast Cancer. Cell Metab. 2018 Jul 3;28(1):87-103.e6. doi: 10.1016/j.cmet.2018.04.022. Epub 2018 May 24. PubMed PMID: 29805099.
3. Kahl, D.J., Hutchings, K.M., Lisabeth, E.M., Haak, A.J., Leipprandt, J.R., Dexheimer, T., Khanna, D., Tsou, P.S., Campbell, P.L., Fox, D.A., Wen, B., Sun, D., Bailie, M., Neubig, R.R. & Larsen, S.D. 5-Aryl-1,3,4-oxadiazol-2-ylthioalkanoic Acids: A Highly Potent New Class of Inhibitors of Rho/Myocardin-Related Transcription Factor (MRTF)/Serum Response Factor (SRF)-Mediated Gene Transcription as Potential Antifibrotic Agents for Scleroderma. J Med Chem 62, 4350-4369 (2019) PMC6590913.
4. Madak, J.T., Cuthbertson, C.R., Miyata, Y., Tamura, S., Petrunak, E.M., Stuckey, J.A., Han, Y., He, M., Sun, D., Showalter, H.D. & Neamati, N. Design, Synthesis, and Biological Evaluation of 4-Quinoline Carboxylic Acids as Inhibitors of Dihydroorotate Dehydrogenase. J Med Chem 61, 5162-5186 (2018)
5. Zhao Y, Bai L, Liu L, McEachern D, Stuckety JA, Meagher JL, Yang CY, Ran X, Zhou B, Hu Y, Li X, Wen B, Zhao T, Li S, Sun D, Wang S. Structure-based discovery of 4-(6-methoxy-2-methyl-4-(quinolin-4-yl)-9H-pyrimido[4,5-b]indol-7-yl)-3,5-dimethylisoxazole (CD161) as a potent and orally bioavailable BET bromodomain inhibitor. J Med Chem. 2017 May 11;60(9):3887-3901. PMID: 28463487
6. Zhao Y, Liu L, Sun W, Lu J, McEachern D, Li X, Yu S, Bernard D, Ochsenbein P, Ferey V, Carry JC, Deschamps JR, Sun D, Wang S. Diastereomeric spirooxindoles as highly potent and efficacious MDM2 inhibitors. J Am Chem Soc. 2013;135(19):7223-34. PMCID: PMC3806051
Instruments application notes from different company to describe the capability of the each instrument. (UM PKMS core own those cutting-edge technology and instrument to perform similar studies.)
- SCIEX Triple Quad 5500 LC-MS/MS --- Highly Sensitive and Robust Quantification Method for Ethinyl Estradiol and Drospirenone in plasma https://sciex.com/Documents/tech%20notes/Highly_Sensitive_and_Robust_Quantification.pdf
- SCIEX Triple Quad 5500 Qtrap system --- In Vivo Metabolic Profiling of Carbamazepine Using the QTRAP® 5500 System and LightSight® Software v2.2 https://sciex.com/Documents/Downloads/Literature/mass-spectrometry-Carbamazepine-profiling-1036610.pdf
- SCIEX X500R QTOF system --- Using MS/MSall with SWATH Acquisition For Forensic Designer Drug Analysis with SCIEX X500R QTOF system and SCIEX OS Software https://sciex.com/Documents/tech%20notes/forensic-designer-drug-analysis.pdf
- SCIEX X500R QTOF system --- Rapid Metabolite Identification using MetabolitePilot™ Software and TripleTOF™ 5600 system https://sciex.com/Documents/Downloads/Literature/Rapid_Metabolite_Identification_using_MetabolitePilot_Software.pdf
- SYNAPT G2-Si HDMS QTOF system --- Targeted High Resolution Quantification with Tof-MRM and HD-MRM http://www.waters.com/webassets/cms/library/docs/720004728EN.pdf
- Waters SYNAPT G2-Si HDMS QTOF system --- Qualitative and Quantitative Metabolite Identification for Verapamil in Rat Plasma Using a UPLC/SYNAPT G2 HDMS Strategy with MSE http://www.waters.com/webassets/cms/library/docs/720003492en.pdf
- Waters SYNAPT G2-Si HDMS QTOF system --- Ion Mobility Separation Coupled With Desorption Electrospray Ionization Mass Spectrometry for High Specificity MS Imaging http://www.waters.com/webassets/cms/library/docs/720005525en.pdf
- Waters SYNAPT G2-Si HDMS QTOF system --- Lipid Visualization and Identification Through Collisional Cross Section Aided Correlation of MS Imaging and Ex Situ MS Data for MS-MS Identification http://www.waters.com/webassets/cms/library/docs/720005354en.pdf
Edited by Bo Wen, 05/2020
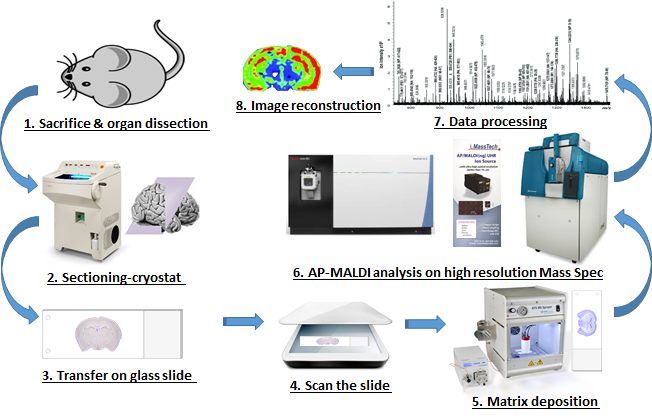
Mass spectrometry imaging (MSI) is an emerging technique that allows molecular visualization of the distribution of drugs, lipids, peptides, proteins and metabolites in a two-dimensional space directly in biological tissues. MSI allows unlabeled drug compounds and drug metabolites to be detected and identified and quantified according to their mass-to-charge ratios (m/z) at high resolution in complex tissue environments. This technique has better specificity and provides the possibility to combine histological data with MS ones and to visualize simultaneously the distribution drugs or biomarkers in relation to tumor heterogeneity, provide new understandings of the dynamic processes impacting drug uptake and metabolism at the local sites targeted by therapy.
We have a state of the art MALDI/DESI SYNAPT G2-Si High Definition Mass Spectrometry (Waters®) in our Pharmacokinetics and Mass Spectrometry Core Facility (PKMS). Using the MALDI and DESI ion source together with T-Wave ion mobility technology, our experienced team of mass spectrometry imagers can provide distribution profiling information for a wide range of sample types, such as tissue specimens, industrial and biological materials, horticultural specimens, and microbiological populations. We offer MSI sample preparation, including cryo-sectioning and microtome techniques, histological staining, and tissue microscopy. We can also combine imaging data with data acquired by different analytical techniques to deliver more comprehensive results. Our team works with you to identify MSI approaches to address your application requirements.
- Drug development
- Pharmacokinetics: Drug and Metabolite Distribution, 2D and 3D label free Imaging
- Disease characterization and biomarker investigations
- Carcinomas
- Inter-and intra-tumor heterogeneity
- Inflammatory diseases
- Fibrosis
- Bacterial infections and manifestations
- Neurological diseases
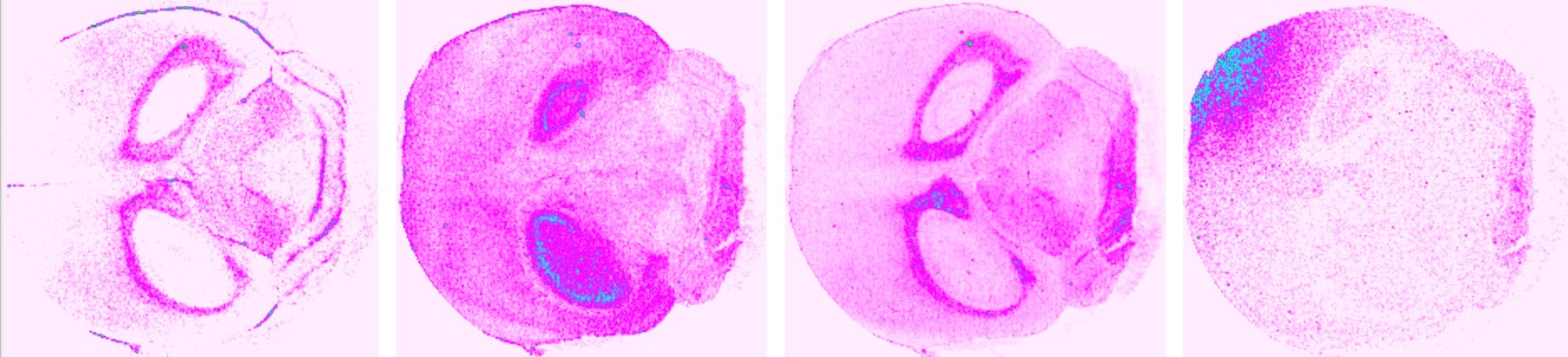
PKMS Case study 1 : small molecular or lipids mass spectrometry imaging on mouse brain tissue section, thousand compounds can be imaged from the same tissue section at same time.
M/Z 253.08 M/Z 314.11 M/Z 495.02 M/Z 831.45
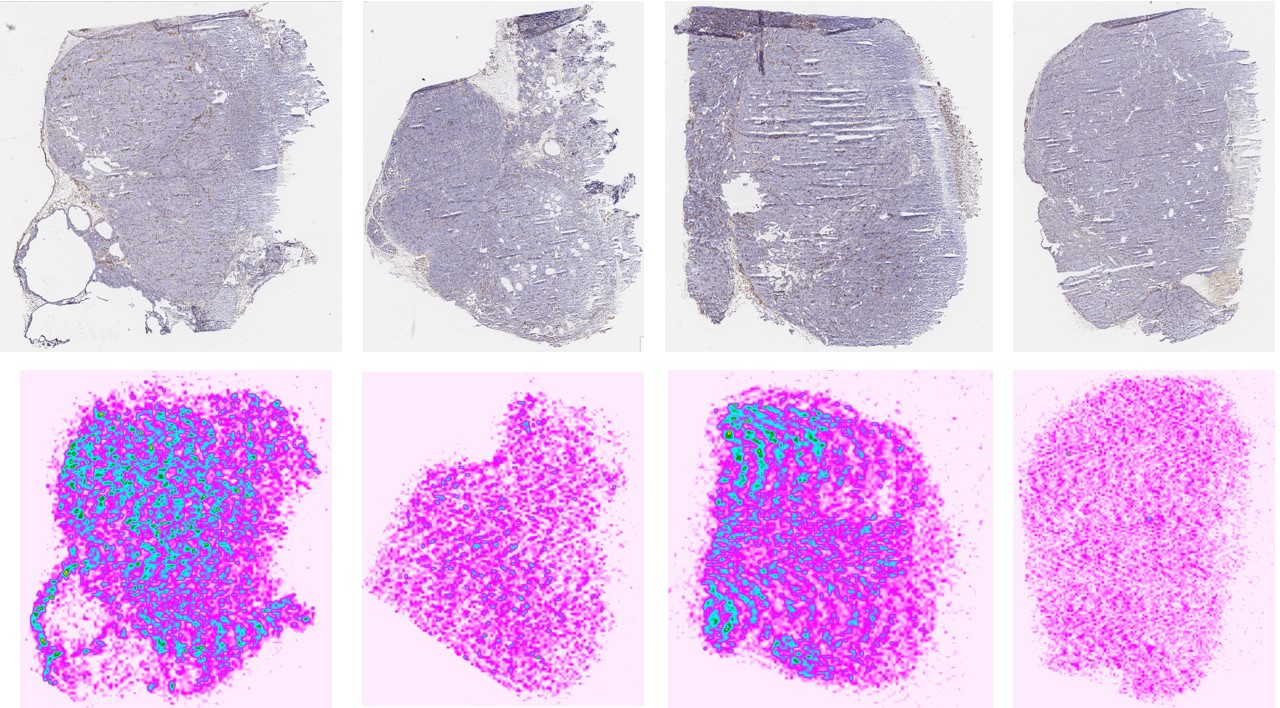
PKMS Case study 2 : MALDI-Ion mobility mass spectrometry imaging for paclitaxel ditribution in solid tumor tissue. https://pharmacy.umich.edu/pkcore/Conference%20Poster
Instruments application notes from Waters® to describe the capability of the SYNAPT G2-Si High Definition Mass Spectrometry.
- Biomarker Discovery Directly from Tissue Xenograph Using High Definition Imaging MALDI Combined with Multivariate Analysis. http://www.waters.com/webassets/cms/library/docs/720004873en.pdf
- Data Independent MALDI Imaging HDMSE for Visualization and Identification of Lipids Directly from a Single Tissue Section. http://www.waters.com/webassets/cms/library/docs/720004471en.pdf
- Distribution of Biomarkers of Interest in Rat Brain Tissues Using High Definition MALDI Imaging. http://www.waters.com/webassets/cms/library/docs/720004135en.pdf
- MALDI Imaging of Distribution of Xanthohumol and Its Metabolites in Rat Tissues. http://www.waters.com/webassets/cms/library/docs/720003707en.pdf
- Successful Application for Distribution Imaging of Chloroquine Ocular Tissue in Pigmented Rat Using MALDI-Imaging Quadrupole Time-of-Flight Mass Spectrometry. http://www.waters.com/waters/library.htm?locale=en_SE&cid=10010845&lid=10180348
- MALDI-Ion Mobility Separation-Mass Spectrometry Imaging of Glucose-Regulated Protein 78 kDa (Grp78) in Human Formalin-Fixed, Paraffin-Embedded Pancreatic Adenocarcinoma Tissue Sections. https://pubs.acs.org/doi/abs/10.1021/pr900522m
- Tissue Imaging of Pharmaceuticals by Ion Mobility Mass Spectrometry. http://www.waters.com/webassets/cms/library/docs/720003216en.pdf
- Direct Tissue Imaging and Characterization of Phospholipids Using MALDI SYNAPT HDMS System. http://www.waters.com/webassets/cms/library/docs/720002444en.pdf
- Localizing Diazepam and its Metabolite in Rat Brain Tissue by Imaging Mass Spectrometry using MALDI Q-Tof Premier MS. http://www.waters.com/webassets/cms/library/docs/720002447en.pdf
- Matrix-Assisted Laser Desorption/Ionization-Ion Mobility Separation-Mass Spectrometry Imaging of Vinblastine in Whole Body Tissue Sections. https://pubs.acs.org/doi/abs/10.1021/ac8015467
Edited by Bo Wen, 05/2020
The use of bioanalytical technologies such as quantitative liquid chromatography-mass spectrometry (LC-MS/MS) has been accepted as the gold-standard technology to support PK and drug metabolism studies. It comes significant improvements in assay sensitivity, selective and specificity, plus the potential to reduce assay time per samples, and the detection of compounds of interest in different matrices, no matter how complex - including plasma, blood, serum, urine, faces, spinal fluid, skin, muscle, artery, myocardium, liver and kidney, and a variety of tumor types. Below is list of drug that have been tested in our core with an validated LC-MS assay.
| Compound Name | Classification | Transition Ions ( M/Z) | Limit of Detection |
| Acalabrutinib | Bruton's tyrosine kinase inhibitor | 466.0→372.3 | 0.2 ng/mL in rat plasma |
| Adenosine | Nucleoside | 268.0→136.0 | 5 ng/ml in plasma |
| Afatinib | Tyrosine kinase inhibitor | 486.1→370.9 | 0.5 ng/mL in human plasma |
| Amifostine | Chemoprotective agent | HRMS | HRMS |
| Aminocaproic Acid | Clotting promoter | 132.1→95.9 | 10 ng/mL in human plasma |
| Amlexanox | Anti-inflammatory agent | 297.1→253.2 | 1 ng/mL in mice serum |
| Apcin | Cell cycle inhibitor | 438.1→224.0 | 2 ng/mL in mice plasma |
| Apixaban | Anticoagulant | 460.2→443.2 | 1 ng/mL in human plasma |
| Arachidonoyl Glycine | Lipids | 362.5→278.0 | 1 ng/mL in acetonitrile |
| Aripiprazole | Atypical antipsychotic | 448.2→285.2 | 1 ng/mL in plasma or brain |
| Artemether | Antimalarial | 316.2→267.2 | 2 ng/mL in human plasma |
| Azithromycin | Antibiotic | 479.5→591.4 | 1 ng/mL in rat blood or brain |
| Buprenorphine | Narcotic | 468.1→396.2 | 0.1 ng/mL in mice plasma |
| Bupropion | Antidepressant | 240.0→184.0 | 0.5 ng/mL in human plasma |
| Carprofen | Nonsteroidal anti-inflammatory drug | 272.0→228.1 | 5 ng/mL in mouse plasma |
| Cefaclor | Second-generation cephalosporin antibiotic | 368.0→174.0 | 2 ng/mL in human plasma |
| Cefazolin | Antibiotic | 455.2→323.0 | 10 ng/mL in human plasma |
| Chlorambucil | Chemotherapy | 303.9→192.0 | 50 ng/mL in mouse plasma |
| Cholesterol | Sterol | 369.3→369.3 | 10 ng/mL in solution |
| Cisplatin | Chemotherapy | 492.0→426.0 | 1 ng/ml in urine |
| Clofazimine | substituted iminophenazine dye | 473.0→431.1 | 1 ng/ml in mouse serum |
| Concanamycin A | plecomacrolide antibiotic | 888.8→515.2 | 0.1 ng/mL in mouse plasma |
| Cyclophosphamide | Chemotherapy | 261.0→140.0 | 1 ng/mL in human plasma |
| Dabrafenib | Kinase inhibitors | 520.0→343.1 | 1 ng/ml in mouse plasma |
| Dacomitinib | multi-kinase receptor inhibitor | 470.0→385.0 | 1 ng/mL in rat plasma |
| Daptomycin | Antibiotic | 811.3→ 341.4 | 1 ng/mL in feces |
| Dasatinib | Chemotherapy | 488.3→ 401.2 | 1 ng/mL in plasma |
| Deoxyuridine | Antiviral | 237.1→137.0 | 5 ng/ml in plasma |
| Dexamethasone | Corticosteroid | 393.0→373.0 | 2.5 ng/mL in plasma |
| Docetaxel | Chemotherapy | 808.4→225.9 | 1 ng/mL in plasma |
| Doxorubicin | Chemotherapy | 544.0→361.0 | 2 ng/mL in plasma |
| Doxycycline | Antibiotic | 445.2→ 321.0 | 2.5 ng/mL in plasma |
| Echinomycin | Antibiotic | 1101.1→1053.2 | 0.1 ng/mL in plasma |
| Eliglustat | Gastrointestinal system drug | 405.1→316.2 | 1 ng/mL in plasma |
| Entrectinib | Chemotherapy | 561.0→302.1 | 1 ng/mL in plasma |
| Ertapenem | Antibiotic | 476.1→346.1 | 100 ng/mL in plasma |
| Etonogestrel | Contraceptive drug | 325.2→257.2 | 0.25 ng/mL in plasma |
| Everolimus | Immunosuppressive agent | 980.2→389.1 | 1 ng/mL in plasma |
| Fedratinib | Inhibitor of myelfobrosis | 525.1→525.1 | 1 ng/mL in plasma |
| Fentanyl | Narcotic | 337.2→188.2 | 1 ng/mL in plasma |
| Ferioxamine | Supply of iron | 614.4→414.4 | 0.48 nM in water |
| Fingolimod | Immunosuppressive drug | 308.4→255.1 | 1 ng/mL in plasma |
| Floxuridine | Chemotherapy | 245.1→155.0 | 2 ng/mL in liver sample |
| Fluticasone | Antiallergy medicine | 501.0→293.2 | 3 ng/mL in plasma |
| Fluticasone Propionate | Steroid and Decongestant | 501.2→293.2 | 1 ng/mL in plasma |
| Fulvestrant | Chemotherapy | 605.2→427.4 | 0.1 ng/mL in plasma |
| Gallein | Chemotherapy | 365.1→189.0 | 5 ng/mL in plasma |
| Glucoraphanin | Supplement | 438.0→196.0 | 2 ng/mL in urine |
| Ibrutinib | Chemotherapy | 441.1→138.1 | 1 ng/mL in plasma |
| Ibuprofen | Nonsteroidal anti-Inflammatory drug | 205.1→125.0 | 5 ng/mL in plasma |
| Imatinib | Chemotherapy | 372.5 → 176.3 | 0.5 ng/mL in plasma |
| Larotrectinib | Chemotherapy | 429.0 → 342.0 | 1 ng/mL in plasma |
| Leuprolide | Hormones | 605.5 → 221 | 0.1 ng/mL in plasma |
| Mesalamine | Nonsteroidal anti-Inflammatory drug | 152.0 → 108.0 | 1 ng/mL in plasma |
| Metformin | Diabetes medication | 130.1 → 71.1 | 5 ng/mL in plasma |
| Metoprolol | Beta blocker | 268.2→116.2 | 1 ng/mL in human serum |
| metronidazole | Antibiotic | 172.1→128 | 10 ng/mL in tissue |
| Mianserin | Antidepressant | 266.0→209.0 | 1 ng/mL in plasma |
| Nalbuphine | Narcotic | 358.2→340.2 | 0.5 ng/mL in mouse plasma |
| Naloxone | Narcotic | 327.6→309.6 | 0.5 ng/mL in monkey plasma |
| Neratinib | Chemotherapy | 557.1→512.0 | 1 ng/mL in plasma |
| Nilotinib | Chemotherapy | 530.1→307.2 | 1 ng/mL in mouse plasma |
| Olaparib | Chemotherapy | 435.3→281.2 | 1 ng/mL in mouse plasma |
| Osimertinib | Chemotherapy | 500.2→71.8 | 1 ng/mL in plasma |
| oxalate | nature compound | HRMS | 1 ug/mL in plasma |
| Paclitaxel | Chemotherapy | 854.4→286.1 | 1 ng/mL in human plasma |
| Panobinostat | Chemotherapy | 350.2→158.1 | 1 ng/mL in mouse plasma |
| Paroxetine | Antidepressant | 330.1→192.2 | 1 ng/mL in mouse plasma |
| Pazopanib | Chemotherapy | 438.1→357.2 | 1 ng/mL in human plasma |
| Pemetrexed | Chemotherapy | 428.3→281.1 | 25 pg/mL in urine |
| Piperacillin | Antibiotic | 518.0→359.0 | 5 ng/mL in plasma |
| Pomalidomide | Immunomodulatory drug | 274.02→201.00 | 0.1 ng/mL in mouse plasma |
| Ponatinib | Chemotherapy | 533.3→231.9 | 1 ng/mL in mouse plasma and brain |
| Propranolol Hydrochloride | Beta-blocker | 260.3→260.3 | 5 nM for dissolution studies |
| Rafarm | Iron supplement | Chelatable iron is detected by HPLC-UV | 15.6 µM in saline and rat serum |
| Selumetinib | Chemotherapy | 459.0→301.1 | 10 ng/mL in rat plasma |
| Simvastatin | Antilipemic agent | 436.3→285.2 | 0.1 ng/mL in mouse plasma and liver |
| Sirolimus | Immunosuppressive drug | 931.5→864.6 | 0.5 ng/mL in human blood |
| Solifenacin | Bladder relaxant | 363→193 | 0.5 ng/mL in human plasma |
| sorafenib | Chemotherapy | 465→252 | 10 ng/mL in human plasma |
| Sulfadiazine | Antibiotic | 251.1→156.0 | 400 µM for dissolution studies |
| Sulforaphane | Supplement | 178 → 113.6 | 1 ng/mL in rat plasma |
| Tacrolimus | Immunosuppressive drug | 821.4→768.5 | 0.5 ng/mL in human blood |
| Tamiflu | Antiviral drug | 313.2→166.1 | 0.5 ng/mL in human plasma |
| Tamoxifen | Chemotherapy | 372.1→72.0 | 1 ng/mL in mouse plasma and tissues |
| Tazobactam | Antibiotic | 299.0→138/254.9 | 0.25 µg/mL in human plasma |
| Thymoquinone | Chemotherapy | HPLC-UV | |
| Tofacitinib | Antineoplastic | 313.1→173.1 | 1 ng/mL in mouse plasma |
| Trametinib | Chemotherapy | 616→491.0 | 1 ng/mL in mouse plasma |
| Triamcinolone Acetonide | Steroid | 435→415 | 10.0 pg/mL in rabit plasma |
| Valproic Acid | Anticonvulsant | 143.0→143.0 | 5 µg/mL in human plasma |
| Vancomycin | Antibiotic | 725.6 → 100.2 | 0.05 ng/mL in human plasma |
| Varenicline | Smoking Cessation Agent | 212.1 → 169.1 | 0.1 ng/mL in human plasma |
| Vemurafenib | Chemotherapy | 490.1→383.1 | 100 ng/mL in mouse plasma |
| Venofer | Iron supplement | Chelatable iron is detected by HPLC-UV | 15.6 µM in saline and rat serum |
| Withaferin A | Supplement | 471.1→281 | 2 ng/mL in rat plasma and tissues |
| Zanamivir | Antiviral drug | 333→60 | 1 ng/mL in human plasma |
Edited by Bo Wen, 06/09/20
Pharmacokinetics (PK) is the study of the time course of drug’s absorption, distribution, metabolism, and excretion (ADME), what the body interact with a drug, related to the movement of drug into, pass, and out of the body. In order for a drug to be effective in the body, enough of the active form must reach the target to elicit the desired effect. The compound with the best activity against the target enzyme or receptor may have very poor ADME characteristics such as metabolic instability or poor intestinal absorption. When choosing a lead candidate from multiple compounds, the activity of the drug in the efficacy screen must be balanced with acceptable ADME and stability characteristics. Pharmacokinetic studies with mass spectrometry bioanalysis can provide the quantitative information about the half-life of the administrated compound from in vitro and in vivo test, and more useful information of how quickly it can be metabolized or excreted by enzymatically or nonenzymatically. To track the drug accumulation and distribution in organism from in vivo PK study, the mass spectrometry imaging and quantification can come up with innovative solutions to quantify dynamic drug exposures in all targeted tissues, or using artificial intelligence modeling to predict the drug exposure in disease or normal organs.
Capture PK.JPG
Mass spectrometry imaging (MSI) is an emerging technique that allows molecular visualization of the distribution of drugs, lipids, peptides, proteins and metabolites in a two-dimensional space directly in biological tissues. MSI allows unlabeled drug compounds and drug metabolites to be detected and identified and quantified according to their mass-to-charge ratios (m/z) at high resolution in complex tissue environments. This technique has better specificity and provides the possibility to combine histological data with MS ones and to visualize simultaneously the distribution drugs or biomarkers in relation to tumor heterogeneity, provide new understandings of the dynamic processes impacting drug uptake and metabolism at the local sites targeted by therapy.
UM PKMS MSI Workflow.png
The use of bioanalytical technologies such as quantitative liquid chromatography-mass spectrometry (LC-MS/MS) has been accepted as the gold-standard technology to support PK and drug metabolism studies. It comes significant improvements in assay sensitivity, selective and specificity, plus the potential to reduce assay time per samples, and the detection of compounds of interest in different matrices, no matter how complex - including plasma, blood, serum, urine, faces, spinal fluid, skin, muscle, artery, myocardium, liver and kidney, and a variety of tumor types. Below is list of drug that have been tested in our core with an validated LC-MS assay.
| Compound Name | Classification | Transition Ions ( M/Z) | Limit of Detection |
| Acalabrutinib | Bruton's tyrosine kinase inhibitor | 466.0→372.3 | 0.2 ng/mL in rat plasma |
| Adenosine | Nucleoside | 268.0→136.0 | 5 ng/ml in plasma |
| Afatinib | Tyrosine kinase inhibitor | 486.1→370.9 | 0.5 ng/mL in human plasma |
| Amifostine | Chemoprotective agent | HRMS | HRMS |
| Aminocaproic Acid | Clotting promoter | 132.1→95.9 | 10 ng/mL in human plasma |
| Amlexanox | Anti-inflammatory agent | 297.1→253.2 | 1 ng/mL in mice serum |
| Apcin | Cell cycle inhibitor | 438.1→224.0 | 2 ng/mL in mice plasma |
| Apixaban | Anticoagulant | 460.2→443.2 | 1 ng/mL in human plasma |
| Arachidonoyl Glycine | Lipids | 362.5→278.0 | 1 ng/mL in acetonitrile |
| Aripiprazole | Atypical antipsychotic | 448.2→285.2 | 1 ng/mL in plasma or brain |
| Artemether | Antimalarial | 316.2→267.2 | 2 ng/mL in human plasma |
| Azithromycin | Antibiotic | 479.5→591.4 | 1 ng/mL in rat blood or brain |
| Buprenorphine | Narcotic | 468.1→396.2 | 0.1 ng/mL in mice plasma |
| Bupropion | Antidepressant | 240.0→184.0 | 0.5 ng/mL in human plasma |
| Carprofen | Nonsteroidal anti-inflammatory drug | 272.0→228.1 | 5 ng/mL in mouse plasma |
| Cefaclor | Second-generation cephalosporin antibiotic | 368.0→174.0 | 2 ng/mL in human plasma |
| Cefazolin | Antibiotic | 455.2→323.0 | 10 ng/mL in human plasma |
| Chlorambucil | Chemotherapy | 303.9→192.0 | 50 ng/mL in mouse plasma |
| Cholesterol | Sterol | 369.3→369.3 | 10 ng/mL in solution |
| Cisplatin | Chemotherapy | 492.0→426.0 | 1 ng/ml in urine |
| Clofazimine | substituted iminophenazine dye | 473.0→431.1 | 1 ng/ml in mouse serum |
| Concanamycin A | plecomacrolide antibiotic | 888.8→515.2 | 0.1 ng/mL in mouse plasma |
| Cyclophosphamide | Chemotherapy | 261.0→140.0 | 1 ng/mL in human plasma |
| Dabrafenib | Kinase inhibitors | 520.0→343.1 | 1 ng/ml in mouse plasma |
| Dacomitinib | multi-kinase receptor inhibitor | 470.0→385.0 | 1 ng/mL in rat plasma |
| Daptomycin | Antibiotic | 811.3→ 341.4 | 1 ng/mL in feces |
| Dasatinib | Chemotherapy | 488.3→ 401.2 | 1 ng/mL in plasma |
| Deoxyuridine | Antiviral | 237.1→137.0 | 5 ng/ml in plasma |
| Dexamethasone | Corticosteroid | 393.0→373.0 | 2.5 ng/mL in plasma |
| Docetaxel | Chemotherapy | 808.4→225.9 | 1 ng/mL in plasma |
| Doxorubicin | Chemotherapy | 544.0→361.0 | 2 ng/mL in plasma |
| Doxycycline | Antibiotic | 445.2→ 321.0 | 2.5 ng/mL in plasma |
| Echinomycin | Antibiotic | 1101.1→1053.2 | 0.1 ng/mL in plasma |
| Eliglustat | Gastrointestinal system drug | 405.1→316.2 | 1 ng/mL in plasma |
| Entrectinib | Chemotherapy | 561.0→302.1 | 1 ng/mL in plasma |
| Ertapenem | Antibiotic | 476.1→346.1 | 100 ng/mL in plasma |
| Etonogestrel | Contraceptive drug | 325.2→257.2 | 0.25 ng/mL in plasma |
| Everolimus | Immunosuppressive agent | 980.2→389.1 | 1 ng/mL in plasma |
| Fedratinib | Inhibitor of myelfobrosis | 525.1→525.1 | 1 ng/mL in plasma |
| Fentanyl | Narcotic | 337.2→188.2 | 1 ng/mL in plasma |
| Ferioxamine | Supply of iron | 614.4→414.4 | 0.48 nM in water |
| Fingolimod | Immunosuppressive drug | 308.4→255.1 | 1 ng/mL in plasma |
| Floxuridine | Chemotherapy | 245.1→155.0 | 2 ng/mL in liver sample |
| Fluticasone | Antiallergy medicine | 501.0→293.2 | 3 ng/mL in plasma |
| Fluticasone Propionate | Steroid and Decongestant | 501.2→293.2 | 1 ng/mL in plasma |
| Fulvestrant | Chemotherapy | 605.2→427.4 | 0.1 ng/mL in plasma |
| Gallein | Chemotherapy | 365.1→189.0 | 5 ng/mL in plasma |
| Glucoraphanin | Supplement | 438.0→196.0 | 2 ng/mL in urine |
| Ibrutinib | Chemotherapy | 441.1→138.1 | 1 ng/mL in plasma |
| Ibuprofen | Nonsteroidal anti-Inflammatory drug | 205.1→125.0 | 5 ng/mL in plasma |
| Imatinib | Chemotherapy | 372.5 → 176.3 | 0.5 ng/mL in plasma |
| Larotrectinib | Chemotherapy | 429.0 → 342.0 | 1 ng/mL in plasma |
| Leuprolide | Hormones | 605.5 → 221 | 0.1 ng/mL in plasma |
| Mesalamine | Nonsteroidal anti-Inflammatory drug | 152.0 → 108.0 | 1 ng/mL in plasma |
| Metformin | Diabetes medication | 130.1 → 71.1 | 5 ng/mL in plasma |
| Metoprolol | Beta blocker | 268.2→116.2 | 1 ng/mL in human serum |
| metronidazole | Antibiotic | 172.1→128 | 10 ng/mL in tissue |
| Mianserin | Antidepressant | 266.0→209.0 | 1 ng/mL in plasma |
| Nalbuphine | Narcotic | 358.2→340.2 | 0.5 ng/mL in mouse plasma |
| Naloxone | Narcotic | 327.6→309.6 | 0.5 ng/mL in monkey plasma |
| Neratinib | Chemotherapy | 557.1→512.0 | 1 ng/mL in plasma |
| Nilotinib | Chemotherapy | 530.1→307.2 | 1 ng/mL in mouse plasma |
| Olaparib | Chemotherapy | 435.3→281.2 | 1 ng/mL in mouse plasma |
| Osimertinib | Chemotherapy | 500.2→71.8 | 1 ng/mL in plasma |
| oxalate | nature compound | HRMS | 1 ug/mL in plasma |
| Paclitaxel | Chemotherapy | 854.4→286.1 | 1 ng/mL in human plasma |
| Panobinostat | Chemotherapy | 350.2→158.1 | 1 ng/mL in mouse plasma |
| Paroxetine | Antidepressant | 330.1→192.2 | 1 ng/mL in mouse plasma |
| Pazopanib | Chemotherapy | 438.1→357.2 | 1 ng/mL in human plasma |
| Pemetrexed | Chemotherapy | 428.3→281.1 | 25 pg/mL in urine |
| Piperacillin | Antibiotic | 518.0→359.0 | 5 ng/mL in plasma |
| Pomalidomide | Immunomodulatory drug | 274.02→201.00 | 0.1 ng/mL in mouse plasma |
| Ponatinib | Chemotherapy | 533.3→231.9 | 1 ng/mL in mouse plasma and brain |
| Propranolol Hydrochloride | Beta-blocker | 260.3→260.3 | 5 nM for dissolution studies |
| Rafarm | Iron supplement | Chelatable iron is detected by HPLC-UV | 15.6 µM in saline and rat serum |
| Selumetinib | Chemotherapy | 459.0→301.1 | 10 ng/mL in rat plasma |
| Simvastatin | Antilipemic agent | 436.3→285.2 | 0.1 ng/mL in mouse plasma and liver |
| Sirolimus | Immunosuppressive drug | 931.5→864.6 | 0.5 ng/mL in human blood |
| Solifenacin | Bladder relaxant | 363→193 | 0.5 ng/mL in human plasma |
| sorafenib | Chemotherapy | 465→252 | 10 ng/mL in human plasma |
| Sulfadiazine | Antibiotic | 251.1→156.0 | 400 µM for dissolution studies |
| Sulforaphane | Supplement | 178 → 113.6 | 1 ng/mL in rat plasma |
| Tacrolimus | Immunosuppressive drug | 821.4→768.5 | 0.5 ng/mL in human blood |
| Tamiflu | Antiviral drug | 313.2→166.1 | 0.5 ng/mL in human plasma |
| Tamoxifen | Chemotherapy | 372.1→72.0 | 1 ng/mL in mouse plasma and tissues |
| Tazobactam | Antibiotic | 299.0→138/254.9 | 0.25 µg/mL in human plasma |
| Thymoquinone | Chemotherapy | HPLC-UV | |
| Tofacitinib | Antineoplastic | 313.1→173.1 | 1 ng/mL in mouse plasma |
| Trametinib | Chemotherapy | 616→491.0 | 1 ng/mL in mouse plasma |
| Triamcinolone Acetonide | Steroid | 435→415 | 10.0 pg/mL in rabit plasma |
| Valproic Acid | Anticonvulsant | 143.0→143.0 | 5 µg/mL in human plasma |
| Vancomycin | Antibiotic | 725.6 → 100.2 | 0.05 ng/mL in human plasma |
| Varenicline | Smoking Cessation Agent | 212.1 → 169.1 | 0.1 ng/mL in human plasma |
| Vemurafenib | Chemotherapy | 490.1→383.1 | 100 ng/mL in mouse plasma |
| Venofer | Iron supplement | Chelatable iron is detected by HPLC-UV | 15.6 µM in saline and rat serum |
| Withaferin A | Supplement | 471.1→281 | 2 ng/mL in rat plasma and tissues |
| Zanamivir | Antiviral drug | 333→60 | 1 ng/mL in human plasma |
Edited by Bo Wen, 06/09/20
Listing Row
- Academic Departments
- Research
- Academic Departments
- Research in Clinical Pharmacy
- Research in Medicinal Chemistry
- Research in Pharmaceutical Sciences
- Research Cores and Services
- Biointerfaces Institute
- Michigan Drug Discovery
- Michigan Institute for Clinical & Health Research
- Translational Oncology Program
- Faculty Publications
- News
- UM Pharmacy Professor Research Outreach (PRO)
- About the College