
Helical Dimers
Self-association of transmembrane helices
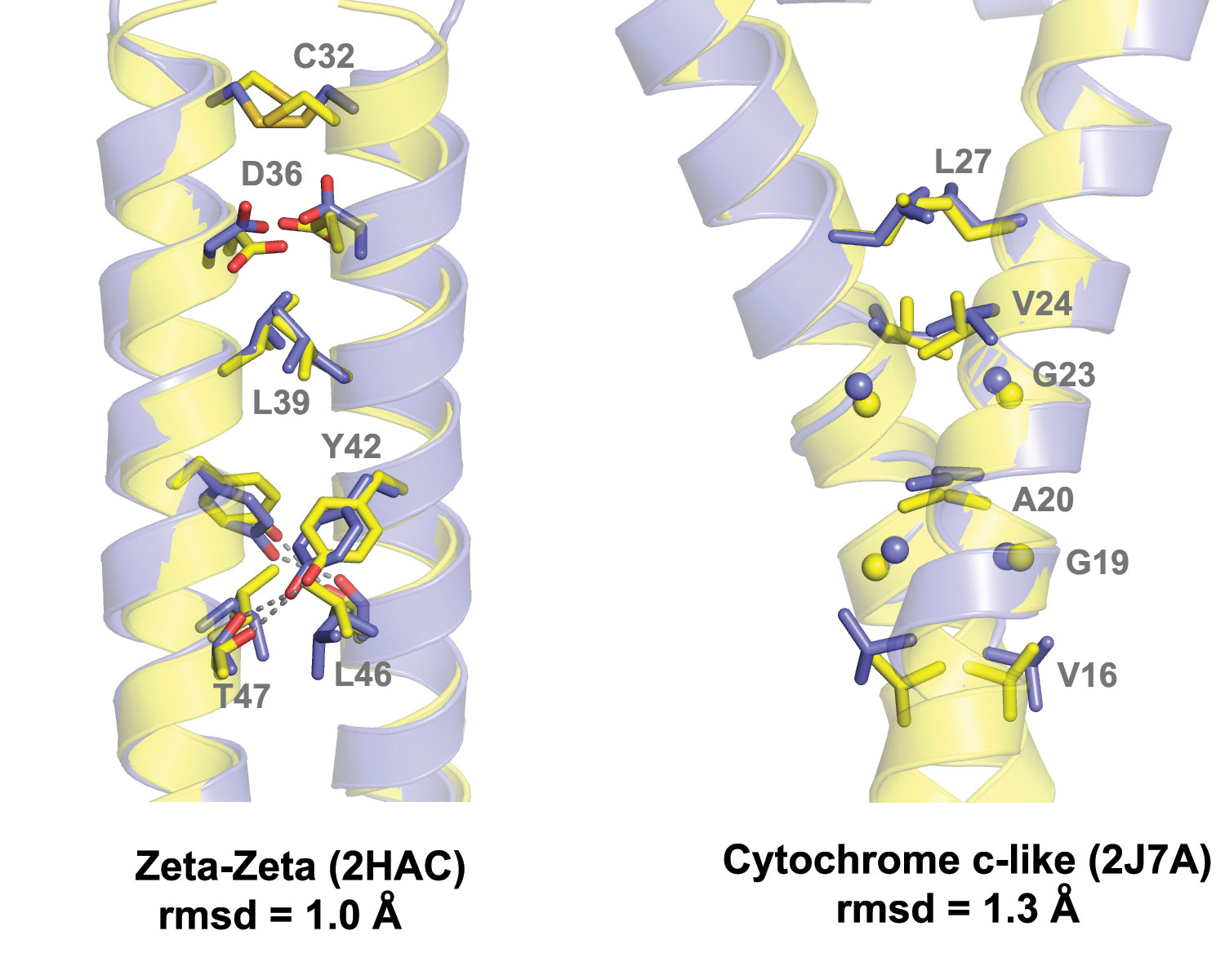 Transmembrane (TM) α-helices of bitopic proteins have a high tendency to form functionally important homo- or hetero-oligomers. Considering the biological significance of bitopic protein oligomers, we have developed the predictive computational method TMDOCK for 3D modeling of TM α-helical dimers in membranes.
Transmembrane (TM) α-helices of bitopic proteins have a high tendency to form functionally important homo- or hetero-oligomers. Considering the biological significance of bitopic protein oligomers, we have developed the predictive computational method TMDOCK for 3D modeling of TM α-helical dimers in membranes.
TMDOCK generates 3D models of dimers by threading a target amino acid sequence through several structural templates, followed by local energy minimization. This is the first method that identifies helix dimerization modes and ranks them based on the calculated free energy of α-helix association. The current version of the TMDOCK web server allows fast and reliable modeling of 3D structures of parallel TM homodimers (Fig.5). After screening of Membranome by TMDOCK, ~2000 potentially biologically relevant TM homodimers were included into the database.


