
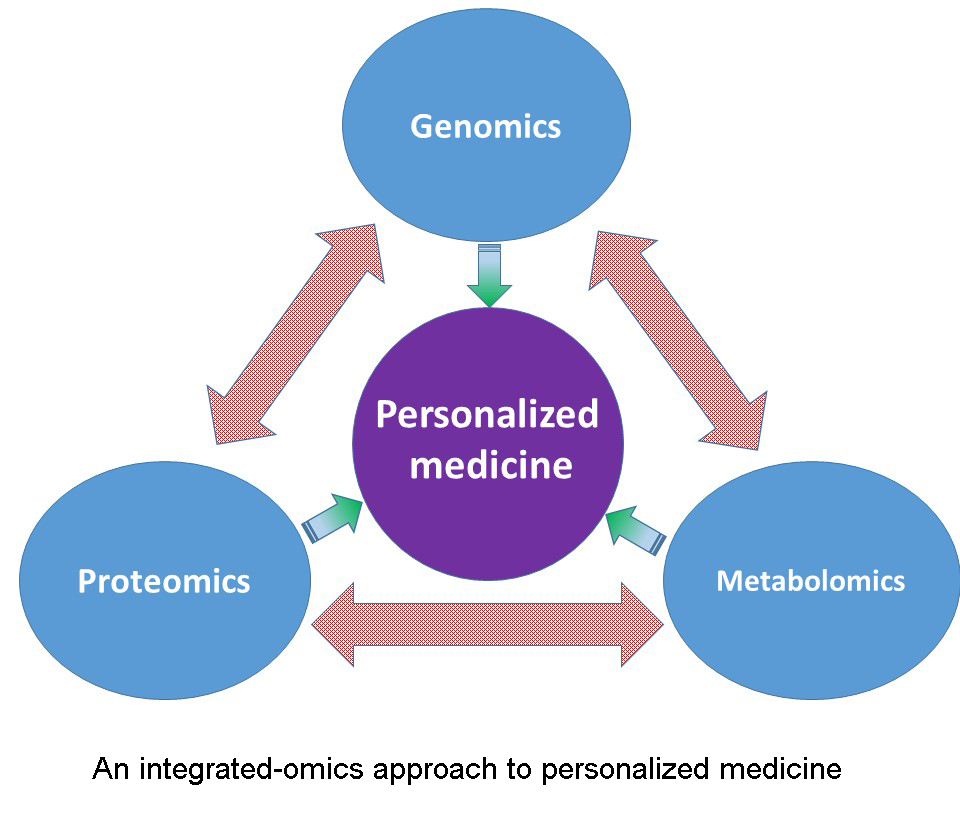
The research in the Zhu lab focuses on identifying genetic and environmental factors that impact the pharmacokinetics and pharmacodynamics of therapeutic agents. The Zhu lab uses a multi-omics approach (e.g., pharmacogenomics and proteomics) to determine genetic variants and protein biomarkers associated with interindividual variability in response to pharmacotherapy. The long-term goal is to develop a multi-omics-based precision pharmacotherapy strategy to enhance the efficacy and safety of drug treatment.
Listing Row
Wednesday, October 9, 2013
Wednesday, October 9, 2013


