
Drug Discovery for Acute Diarrheal Disease - Shigella
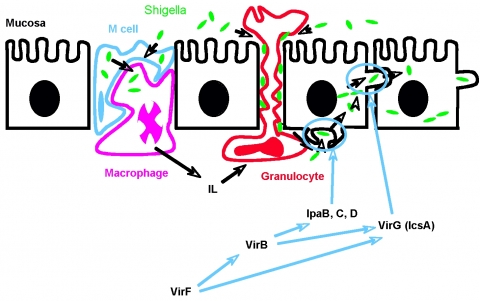 Shigella flexneri is a human enteropathogen that infects ca. 165 million people and claims more than one million lives per year worldwide. Shigella infects the cells of the gastrointestinal tract causing severe dysentery. Absent appropriate antibiotic and electrolyte replacement treatment, shigellosis rapidly leads to dehydration and death. This organism has the ability to travel from cell-to-cell along the intestinal lumen, thereby “hiding” from the host immune system. While shigellosis has been considered to be a disease of the “third world”, like many other highly contagious diseases it is rapidly spreading to developed countries. A recent review of shigellosis in Oregon (Replogle et al., Clin. Infec. Dis. 30 515-9 (2000)) states that of the Shigella strains isolated and studied, 59% were resistant to TMP-SMZ, 63% were resistant to ampicillin, 0.3% were resistant to nalidixic acid and 13% exhibited multi-drug resistance. Clearly, there is a growing need for novel agents against resistant strains of Shigella. Like many pathogens, Shigellaexpresses virulence proteins that are responsible for pathogenicity. Current thinking is that targeting virulence pathways is attractive because drugs that target virulence are expected to exhibit less emergence of drug resistance as there is little selection pressure for resistant strains to out grow wild-type. We are focusing our efforts on VirF, an AraC-type transcriptional regulator that initiates expression of the key shigella virulence genes. We have successfully developed a high-throughput screening assay for VirF activity and identified promising leads from a screen of ~142,000 compounds. Those hits have shown activity in infection model assays performed in collaboration with the Maurelli lab at USUHS in Maryland. In collaboration with the Sherman lab here at UM, we have also conducted a screen of ~20,000 natural product extracts and are in the process of characterizing the hits from that screen.
Shigella flexneri is a human enteropathogen that infects ca. 165 million people and claims more than one million lives per year worldwide. Shigella infects the cells of the gastrointestinal tract causing severe dysentery. Absent appropriate antibiotic and electrolyte replacement treatment, shigellosis rapidly leads to dehydration and death. This organism has the ability to travel from cell-to-cell along the intestinal lumen, thereby “hiding” from the host immune system. While shigellosis has been considered to be a disease of the “third world”, like many other highly contagious diseases it is rapidly spreading to developed countries. A recent review of shigellosis in Oregon (Replogle et al., Clin. Infec. Dis. 30 515-9 (2000)) states that of the Shigella strains isolated and studied, 59% were resistant to TMP-SMZ, 63% were resistant to ampicillin, 0.3% were resistant to nalidixic acid and 13% exhibited multi-drug resistance. Clearly, there is a growing need for novel agents against resistant strains of Shigella. Like many pathogens, Shigellaexpresses virulence proteins that are responsible for pathogenicity. Current thinking is that targeting virulence pathways is attractive because drugs that target virulence are expected to exhibit less emergence of drug resistance as there is little selection pressure for resistant strains to out grow wild-type. We are focusing our efforts on VirF, an AraC-type transcriptional regulator that initiates expression of the key shigella virulence genes. We have successfully developed a high-throughput screening assay for VirF activity and identified promising leads from a screen of ~142,000 compounds. Those hits have shown activity in infection model assays performed in collaboration with the Maurelli lab at USUHS in Maryland. In collaboration with the Sherman lab here at UM, we have also conducted a screen of ~20,000 natural product extracts and are in the process of characterizing the hits from that screen.


