
RNA Base Modification
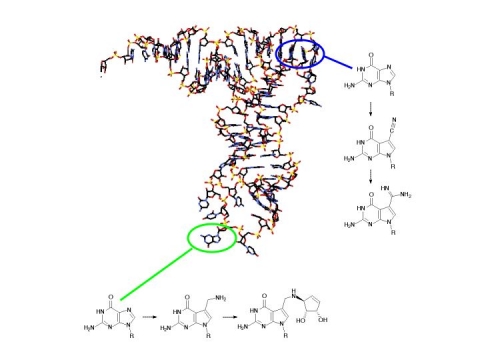 One of our long-standing research interests involves the biosynthesis and physiological roles of modified nucleosides in nucleic acids. Approximately 100 chemically distinct modified nucleosides have been found in nucleic acids, ca. 75% of them in tRNAs. The process of base modification in RNA has many implications in human disease including autoimmune disease, cancer, and viral disease. All tRNAs are post-transcriptionally modified such that anywhere from 5% to 20% (depending upon the species) of the bases contain some covalent modification. These modifications vary from the very simple, such as methylation, to the very elaborate. It has been estimated that 5% of the E. coli genome codes for RNA modifying enzymes, yet relatively little is known of their physiological roles. It remains unclear why tRNAs have so many and such diverse modifications when relatively few are found in other RNA species (although more are being identified). We have done much to explore mechanism, substrate recognition and inhibitor design for the eubacterial tRNA-guanine transglycosylase (TGT), the key enzyme responsible for the queuine modification in the anticodon loop of certain tRNAs. Most recently we have expanded our studies to include the human TGT. These studies are revealing some very interesting evolutionary relationships between the archaeal, eubacterial and eukaryl TGTs and the proposed eukaryl queuine salvage enzyme (eukaryotes do not make queuine, but salvage it from dietary RNA). We are also interested in probing the possibility of base modification in non-coding RNA. We now know that there are ~100,000 non-coding RNA genes in the human genome. It is our hypothesis that these non-coding RNAs contain modified bases that modulate their structure and function.
One of our long-standing research interests involves the biosynthesis and physiological roles of modified nucleosides in nucleic acids. Approximately 100 chemically distinct modified nucleosides have been found in nucleic acids, ca. 75% of them in tRNAs. The process of base modification in RNA has many implications in human disease including autoimmune disease, cancer, and viral disease. All tRNAs are post-transcriptionally modified such that anywhere from 5% to 20% (depending upon the species) of the bases contain some covalent modification. These modifications vary from the very simple, such as methylation, to the very elaborate. It has been estimated that 5% of the E. coli genome codes for RNA modifying enzymes, yet relatively little is known of their physiological roles. It remains unclear why tRNAs have so many and such diverse modifications when relatively few are found in other RNA species (although more are being identified). We have done much to explore mechanism, substrate recognition and inhibitor design for the eubacterial tRNA-guanine transglycosylase (TGT), the key enzyme responsible for the queuine modification in the anticodon loop of certain tRNAs. Most recently we have expanded our studies to include the human TGT. These studies are revealing some very interesting evolutionary relationships between the archaeal, eubacterial and eukaryl TGTs and the proposed eukaryl queuine salvage enzyme (eukaryotes do not make queuine, but salvage it from dietary RNA). We are also interested in probing the possibility of base modification in non-coding RNA. We now know that there are ~100,000 non-coding RNA genes in the human genome. It is our hypothesis that these non-coding RNAs contain modified bases that modulate their structure and function.


